RNA-Seq, ACOBIOM’s expertise to perform RNA Analysis by sequencing

Transcriptomics, or genome-wide expression profiling, aims to catalogue the complete set of RNA transcripts produced by the genome. Transcriptomics provides new biological insight and can be used to determine the structure of genes, their splicing patterns and other post-transcriptional modifications, to detect rare and novel transcripts, and to quantify the changing expression levels of each transcript during development and under different disease conditions.
APPLICATIONS OF RNA-SEQ
Among different approaches, RNA-Seq is a powerful method to identify and quantitatively to decode the entire population of RNAs in a sample, and used in a wide variety of applications. The RNA-Seq is widely used to characterize gene expression patterns associated with the identification of disease-related genes, the analysis of the effects of drugs on tissues, and the discovery of insights in disease pathways. This approach is also used for the :
- Detection of mutation events such as: novel splicing events, gene fusions, SNPs, or other specific coding mutations;
- Detection of rare, or novel transcripts;
- Profiling and comparison of genome-wide expression in different species or otherwise different biological samples.
ADVANTAGES OF RNA-SEQ
RNA-seq provides a more comprehensive view of the transcriptome with a single experiment. RNA-Seq enables to sequence and profile all species of transcripts in the total RNA samples.
RNA-Seq has increased dynamic range and sensitivity. RNA-Seq enables to achieve “digital” transcript expression analysis meaning that expression level data are based on each individual transcript that is sequenced and counted. By increasing the sequencing depth, a potentially unlimited dynamic range can be reached making RNA-Seq an ideal tool for detection rare transcripts.
RNA-Seq provides information of sequence variation in transcripts. RNA-Seq yields a rich data set including information about post-transcriptional mutations and their genomic context. Because RNA-Seq data yields information about how exons are connected, it can reveal sequence variations in the transcripts due to alternative splicing events and provide allele-specific or isoform-specific gene expression information. Moreover, RNA-Seq explore the large family of RNA, with coding (mRNA) and non-coding-RNA like Long non-coding RNA (rRNA, asRNA, pseudogene, …) or small RNA (miRNA, snoRNA, piRNA,…). Additionally, RNA-Seq data is useful for gene mapping functions such as describing the length of UTRs and exon boundaries.
RNA-Seq is not necessarily dependent on any prior sequence knowledge. There is no need for design of probes that must be based on prior sequence or secondary structure information. Therefore, transcriptome profiling in any species is possible which makes this method particularly attractive for non-model species. Additionally, RNA-Seq data can be used to build de novo gene models.
Since RNA-Seq provides absolute values and does not require any calibration with arbitrary standards, results can be compared at any time with other data, even raised by independent laboratories. Once collected, this data can be digitalized and then easily and reliably compared in silico with the growing library of RNA-Seq databases generated for normal and pathological situations in other laboratories around the world
For Instance, ACOBIOM developed the MaRS RNA-Seq database to provide and centralize standardized data used in a wide variety of applications like identifying disease-related genes, analyzing the effects of drugs on tissues or providing insight into disease pathways.
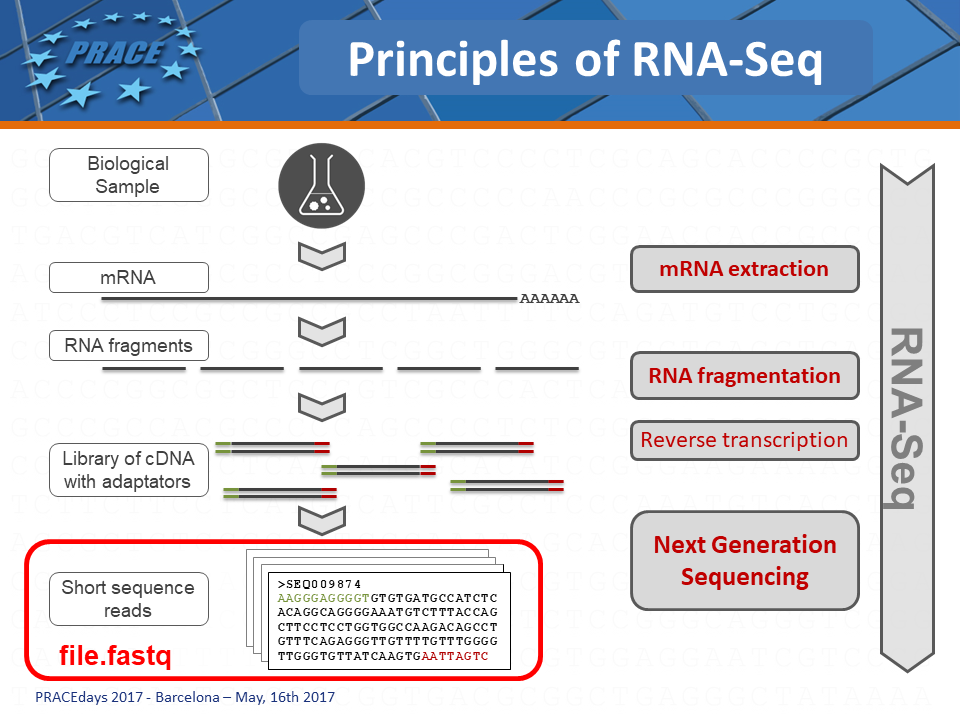
BIOINFORMATICS AND DATA SCIENCE
Dealing with the mass of millions of data generated by RNA-Seq necessitates a catalog of bioinformatics and biostatistics tools. Hence, to analyze the sequencing data generated by this approach, it is necessary to:
- Control the quality of the sequencing data;
- Clean the sequence data;
- Map these data on a reference genome;
- Count the mapped sequences;
- Standardize the data;
- Compare the data with other RNA-Seq profiles;
- Annote and analyze the data;
- Visualize and validate statistically the results.
As these transcriptomic profiles can be easily performed by sequencing, data can be compared to any RNA-Seq profile generated by the scientific community or other laboratories. RNA-Seq can be used in the discovery and the development of new drugs, treatments or diagnostics.
Since its foundation, ACOBIOM investigates RNA molecules and performs RNA analysis by sequencing because genes (DNA) are blueprints, not destiny. Indeed, unlike DNA, which is hardwired at birth and does not change much over our lifetime, RNA is in constant flux, reflecting the overall health on a minute-by-minute basis. RNA is a messenger, carrying instructions from DNA to control protein synthesis and other activities, effectively turning the DNA roadmap into reality.

