Bioinformatics and biostatistics for the analysis of genomic and transcriptomic data
Based on its 20-year experience, Acobiom acquired a unique expertise in the bioinformatic analysis of gene expression and in the identification of RNA biomarkers that are specific to the studied pathology or biological question. The company’s partners or customers will thus be able to benefit from reliable and efficient bioinformatics and biostatistics methods, processes and tools, which have been proven and validated in scientific publications and the development of in-vitro diagnostics.
Bioinformatic and biostatistic analyses adapted to each project
Bioinformatics and biostatistics for sequencing data analysis
Acobiom developed statistical methods and bioinformatic tools dedicated to the analysis and the management of massive amounts of genomic and transcriptomic data generated by sequencing.
Technical team adapts its tools, software and algorithms to the specifications of each analysis and specific treatment.

Acobiom’s team uses customized pipelines and algorithms to:
> the study of the transcriptome, including the expression of genes (encoding or not encoding) or splicing variants,
> the study of the genome, including the analysis of the genome and metagenome, the exome, the methyloma or the degradoma.
Thus, for each of the analyses and each of the stages constituting these analyses, Acobiom selects and adapts the tools and parameters to answer as precisely as possible the biological question studied.
Transcriptome analysis
In the RNA analysis, several types of RNA and processes can be analysed separately or together.
Indeed, the RNA family is a very important family comprising mRNAs (messenger RNAs coding for proteins), and non-coding RNAs, as the miRNA (microRNAs) type or the lncRNA (long non-coding RNAs) type.
The analysis tools and/or parameters used in the analyses are specific to each type of RNA.
Similarly, in the analysis of a process such as splicing, the bioinformatic approach requires a deeper analysis, i.e. it is no longer necessary to consider an RNA as a single entity, but to analyse, identify and count all the exons that compose it.
It should be noted that in humans, nearly 1.2 million exons are now mapped.
The proposed approach to studying these transcriptomes is the RNA-Seq: high throughput sequencing (NGS) of all messenger RNAs present in a given condition.
The transcriptome analysis consists of the following main steps:
a) Quality control and sequence cleaning,
b) Mapping on the reference genome,
c) Counting of the mapped sequences,
d) Standardization and comparison,
e) Sequence annotation,
f) Study of expressed genes: differential analysis, visualization and statistical validation.
Acobiom can also provide upstream the experimental design the most adapted to the research theme in order to obtain the full potential of the data resulting from the associated omics analysis (sequencing, qPCR).
Genome analysis
In the same way as for transcriptome analysis, genome analysis can be approached in different ways: the analysis of coding regions for RNA (Exome), the search for DNA methylation sites (Methyloma) or the search for variants of sequences representative of DNA mutations of somatic (non-hereditary) or constitutional (hereditary) type, characterized by insertions/deletions (InDels) or by mutations of a nucleotide (SNV) on DNA.
Studying an individual’s exome consists of studying the genetic variations of the coding part of the genome in a defined condition
The exome analysis consists of the following main steps:
a) Quality control and sequence cleaning,
b) Mapping on the reference genome,
c) Precise detection of InDels (Insertions/Deletions),
d) Precise detection of SNVs (Single Nucleotide Variants),
e) Sequence annotation,
f) Differential analysis, visualization and statistical validation,
g) Comparison with existing data, such as COSMIC for somatic mutations.
Metagenome analysis and microbiome study (microbial composition)
Metagenomics is the study of all genomes present in a complex environment and in a given condition.
Studying an individual’s microbiota involves identifying the genomes of bacteria or viruses present in a saliva, intestinal or skin sample.
This analysis may consider complete genomes or target specific regions, such as the analysis of 16S RNA (ex V3-V5) in human intestinal microbiota studies.
Microbiome analysis consists of the following main steps:
a) Sequence quality control,
(b) Cleaning and grouping of the representative sequences into OTUs (Operational Taxonomic Units),
c) Population counts,
d) Taxonomic analysis of samples (species detection),
e) Population and diversity analysis (Alpha, Beta diversity),
f) Differential analysis, visualization and statistical validation.
MaRS (Matrix of RNA-Seq): a database of 21,000 Human RNA-Seq profiles
ACOBIOM collected in the same database, called MaRS, 21,000 Human gene expression analyses of RNA-Seq type.
MaRS is focused on the RNA-Seq method, which reflects the expression of genes in a given condition.
Generated by Next Generation Sequencing (NGS), all these profiles were analyzed according to a standardized way on High Performance Computing (Cines HPC machine).
The MaRS matrix thus allows direct comparison of these 21,000 RNA-Seq profiles and the associated transcriptomic data.
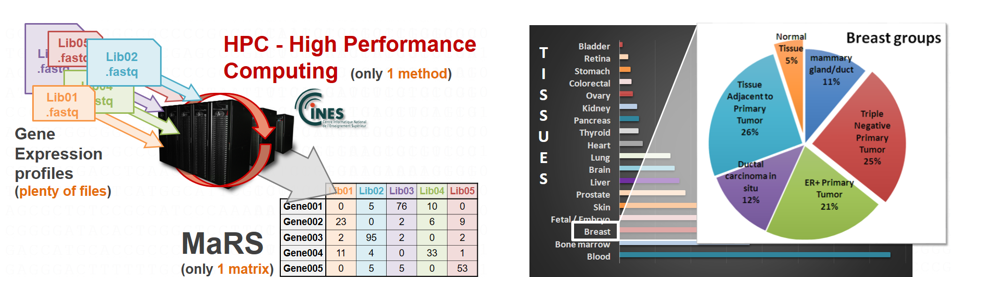
MaRS contains profiles of Human pathologies (cancers, infections, genetic diseases…) and on many organs (blood, breast, lung, liver, pancreas…).
Thus, ACOBIOM uses MaRS to explore and identify new biomarkers, but also to combine these organized and hierarchical data with newly generated data.
In addition, MaRS makes it possible to qualify and/or identify new reference genes (Housekeeping genes) that are just as important as the targeted genes/biomarkers for new PCR analyses (qPCR, RT-qPCR).
The MaRS database is a great internal development tool for ACOBIOM.
The company provides also its partners with access to this database through services and/or scientific collaborations.

